Self Organizing Map Clustering¶
Self Organizing Map (SOM) Clustering is an unsupervised machine learning technique used to visualize and cluster high-dimensional data by projecting it onto a lower-dimensional (typically 2D) grid. SOMs preserve the topological relationships of the input data, making them especially useful for pattern recognition, data exploration, and dimensionality reduction. The algorithm iteratively adjusts a grid of nodes (neurons) to represent the structure of the input data, grouping similar data points together and revealing underlying patterns as presented in the figure below. Once the SOM is trained, each node is associated with a cluster, and new data points can be assigned to the closest node in the grid, allowing for clustering of the data. This process allows for effective clustering of complex datasets, where each node represents a cluster of similar data points. each data point is assigned to the closest node, effectively clustering the data based on its features.
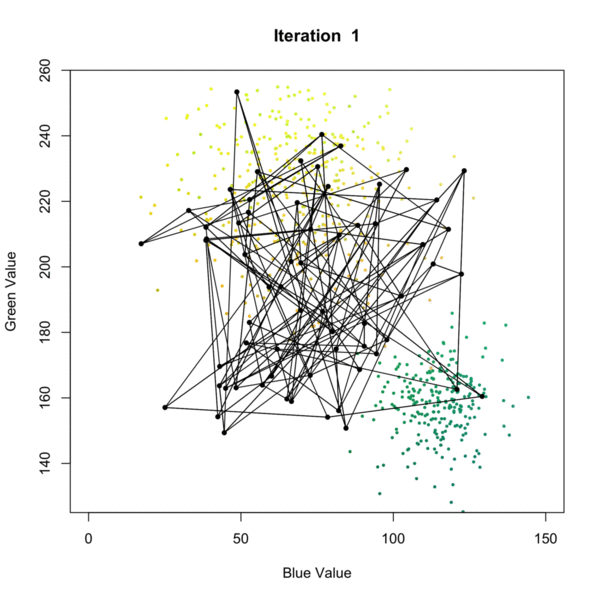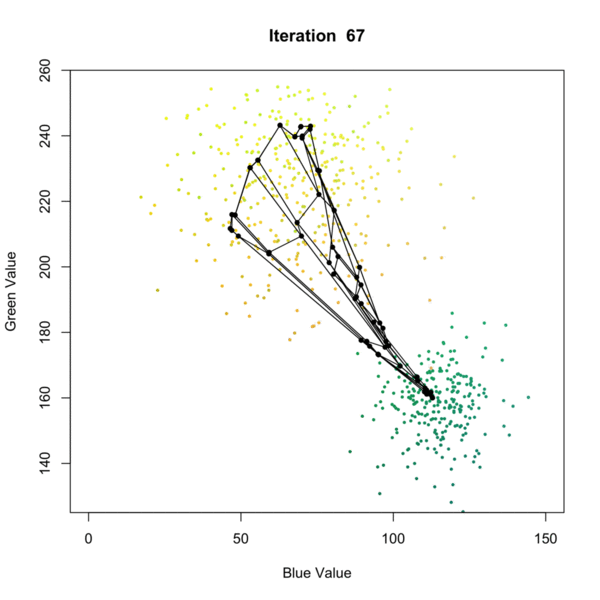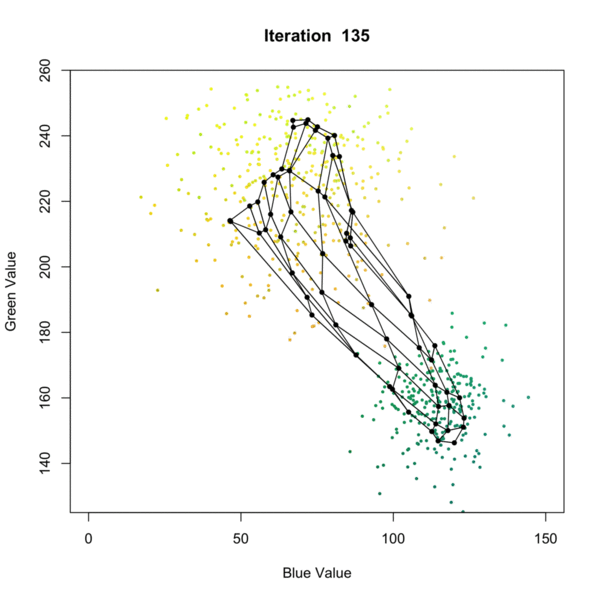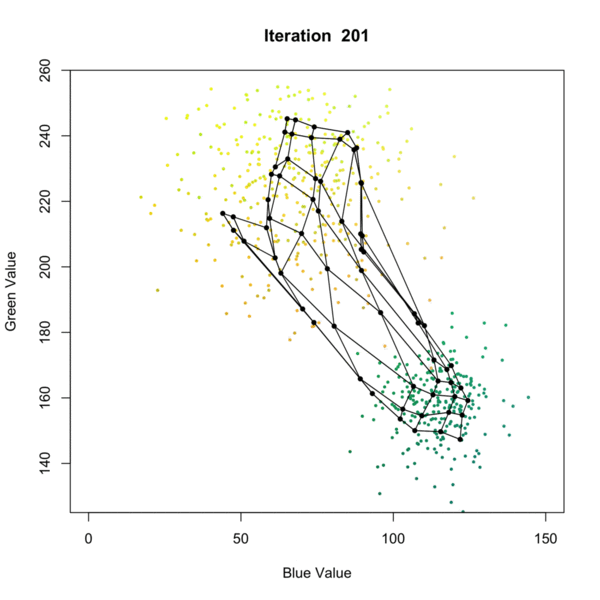
In geosciences, SOM clustering is commonly applied to geochemical, geophysical, or mineralogical datasets to identify distinct zones or patterns. For example, in mineral exploration, SOMs can be used to cluster multi-element geochemical data from soil or rock samples, helping to distinguish between different lithological units, alteration zones, or mineralization styles. This approach aids geologists in targeting areas of interest and understanding the spatial distribution of geochemical signatures.
Interface¶
General parameters¶
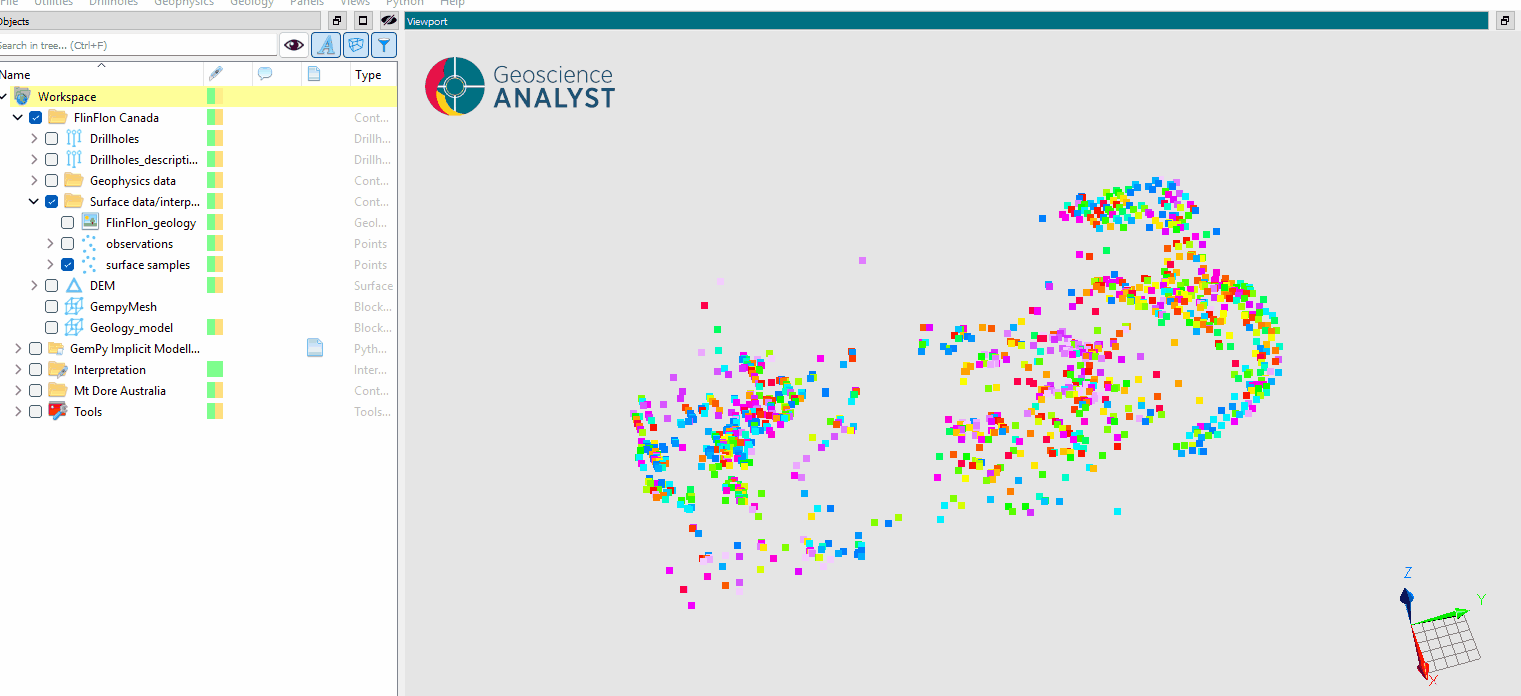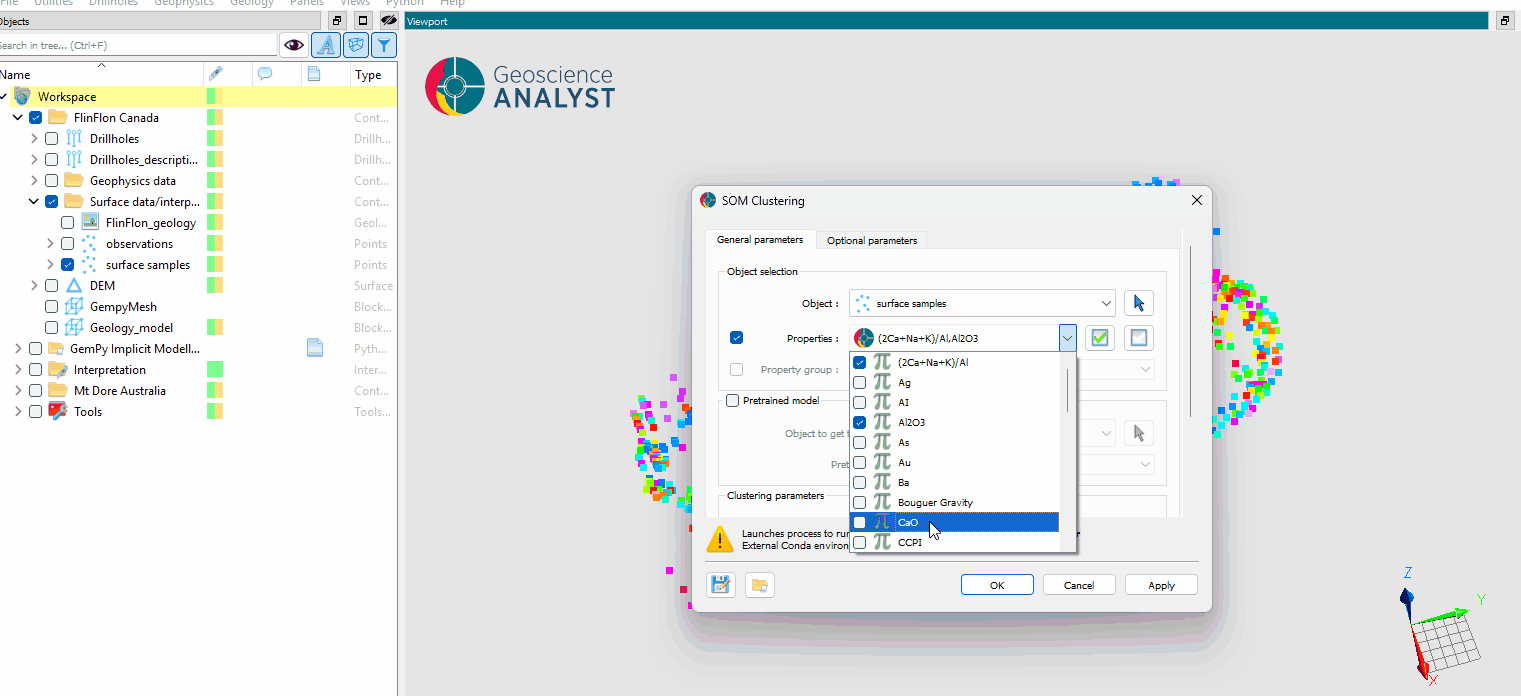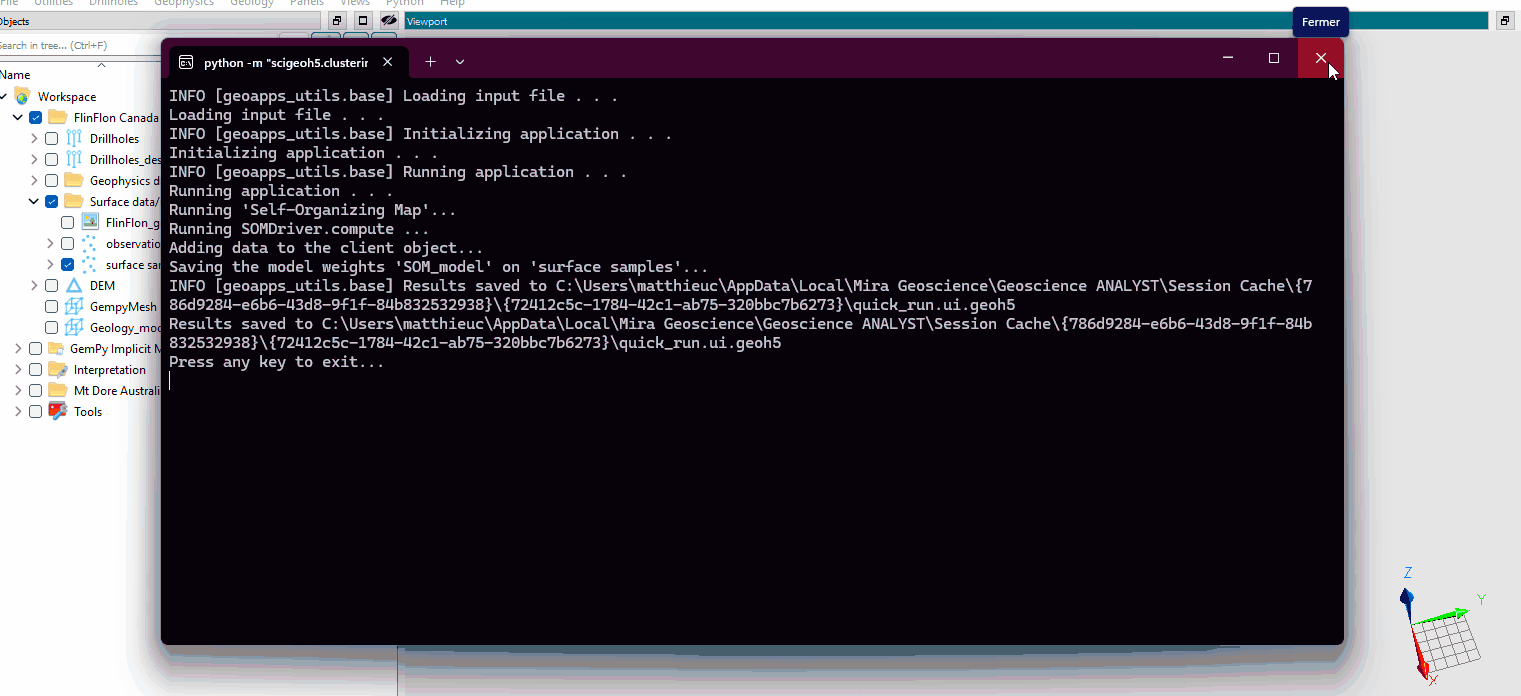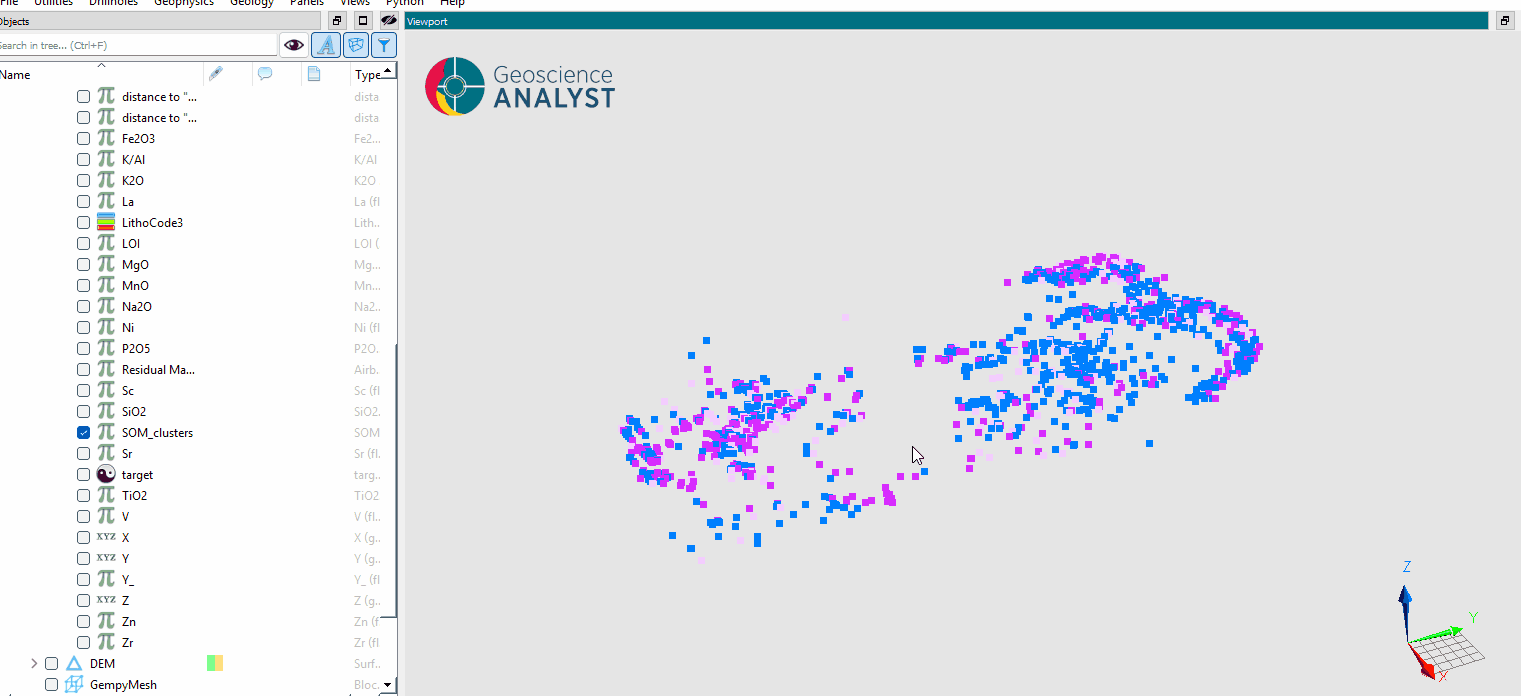
The general parameters controlling the SOM Clustering algorithm are shown in the figure below.
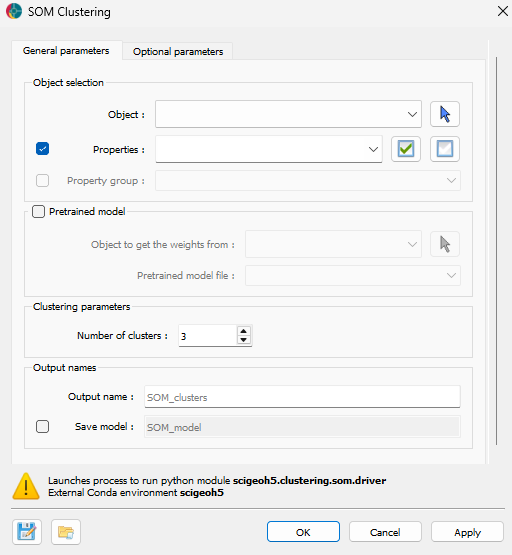
The options are described as follows:
Object Selection¶
Object: The object containing the data to decompose and cluster.
Data: The data properties used for clustering. Users can select multiple
Floatdata properties associated withCellorVertexelements.
Pretrained Model (Optional)¶
Object to get the weights from: Select an object to use pretrained SOM weights.
Pretrained model file: Load a file containing pretrained SOM weights to apply to the data.
Clustering Parameters¶
Number of clusters: The number of clusters to create in the Self-Organizing Map.
Output Names¶
Output name: The name for the resulting clustering data and data group. Default is “SOM_clusters”.
Save model (optional): The name for the output file containing the trained SOM model (saved as a
.pklfile).
Optional Parameters¶
Advanced controls for the SOM Clustering algorithm are available as advanced parameters, as shown in the figure below.
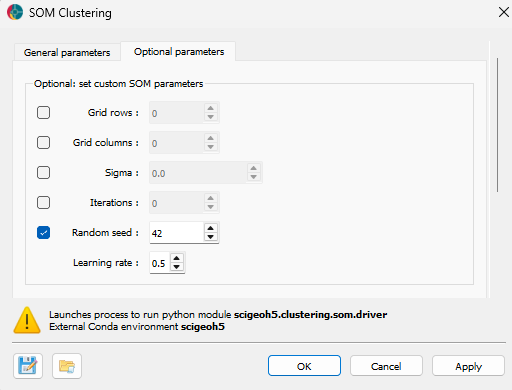
Grid rows (optional): The number of rows in the SOM grid. If not set, the value is determined automatically.
Grid columns (optional): The number of columns in the SOM grid. If not set, the value is determined automatically.
Sigma (optional): The neighborhood radius for updating weights during training. If not set, the value is determined automatically.
Iterations (optional): The number of training iterations. If not set, the value is determined automatically.
Random seed (optional, default = 42): The random seed for reproducibility.
Learning rate (default = 0.5): The initial learning rate for SOM training.
Once the parameters are selected, press OK to run the clustering and obtain results based on the Self-Organizing Map.
Tutorial¶
The following animated image presents a tutorial on how to use the som Clustering application.
Open the application.
Select the object containing the data.
Select the data to be analyzed.
Define the number of clusters.
Run the application.
Inspect the clustering results.