DBSCAN Clustering¶
Density-Based Spatial Clustering of Applications with Noise (DBSCAN) is an unsupervised machine learning algorithm that groups points based on their spatial proximity. It identifies core points that have a minimum number of neighbors within a given “distance”, expanding clusters from these core points while treating isolated points as noise. The “distance” refers to a measure of similarity or difference between data points based on their values across multiple properties. It quantifies how close or far apart two points are in terms of their characteristics, helping to group similar data together.
Interface¶
General parameters¶
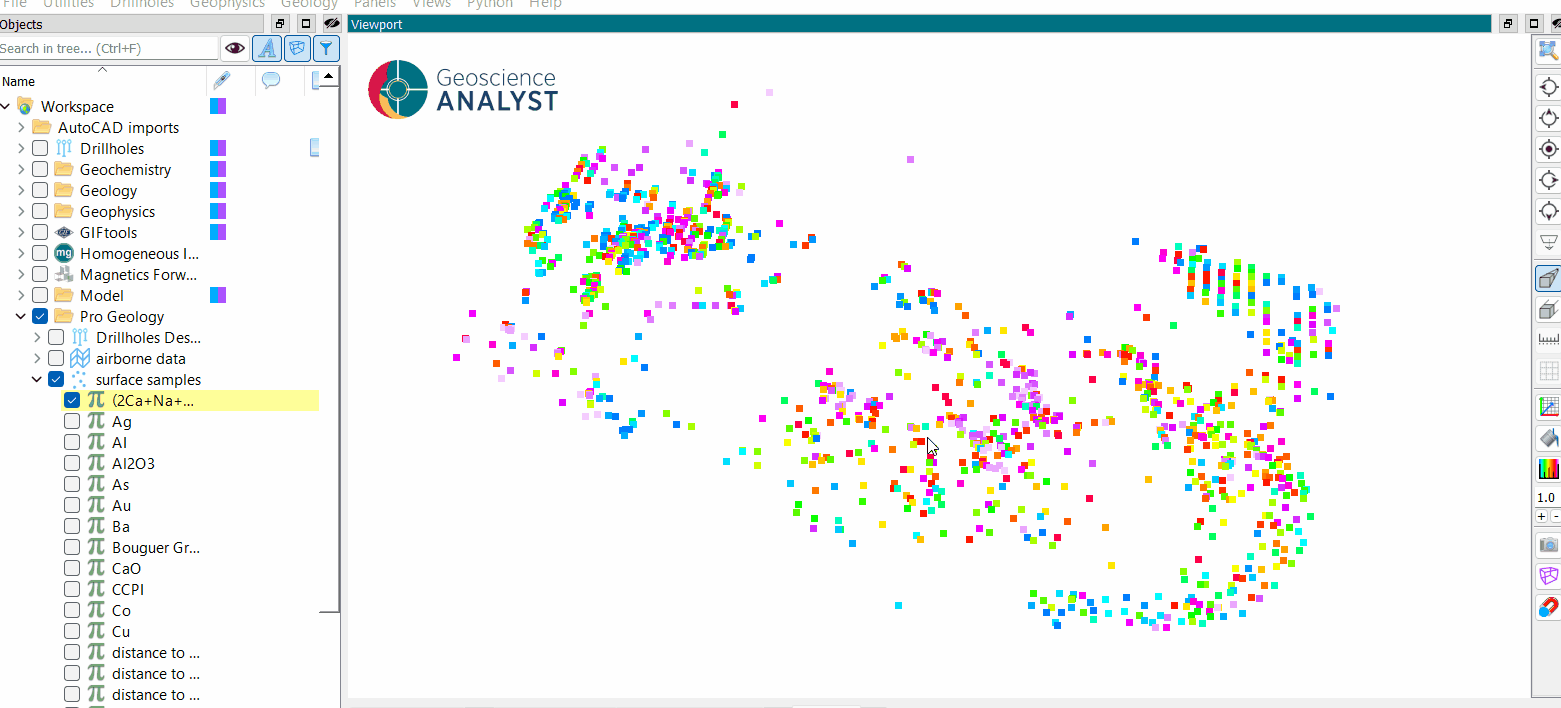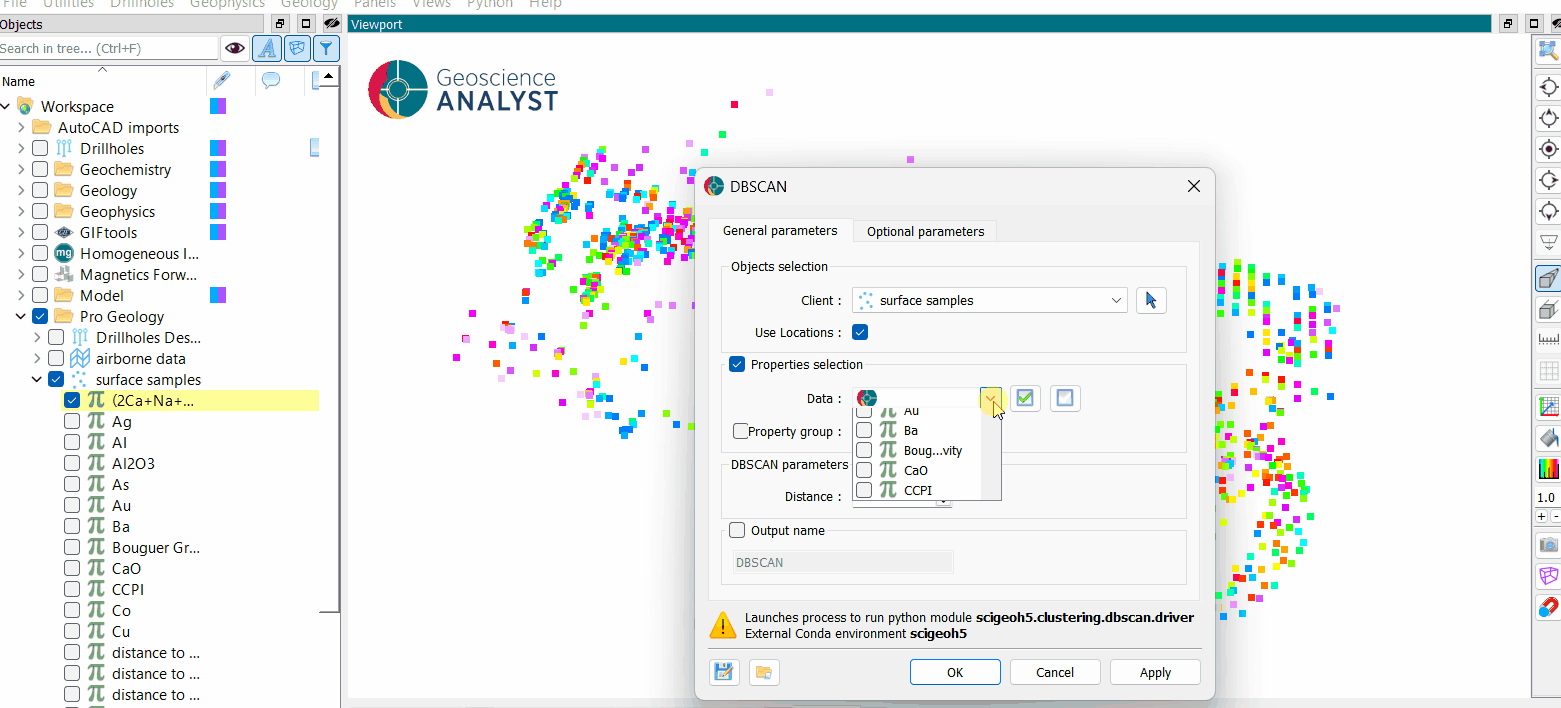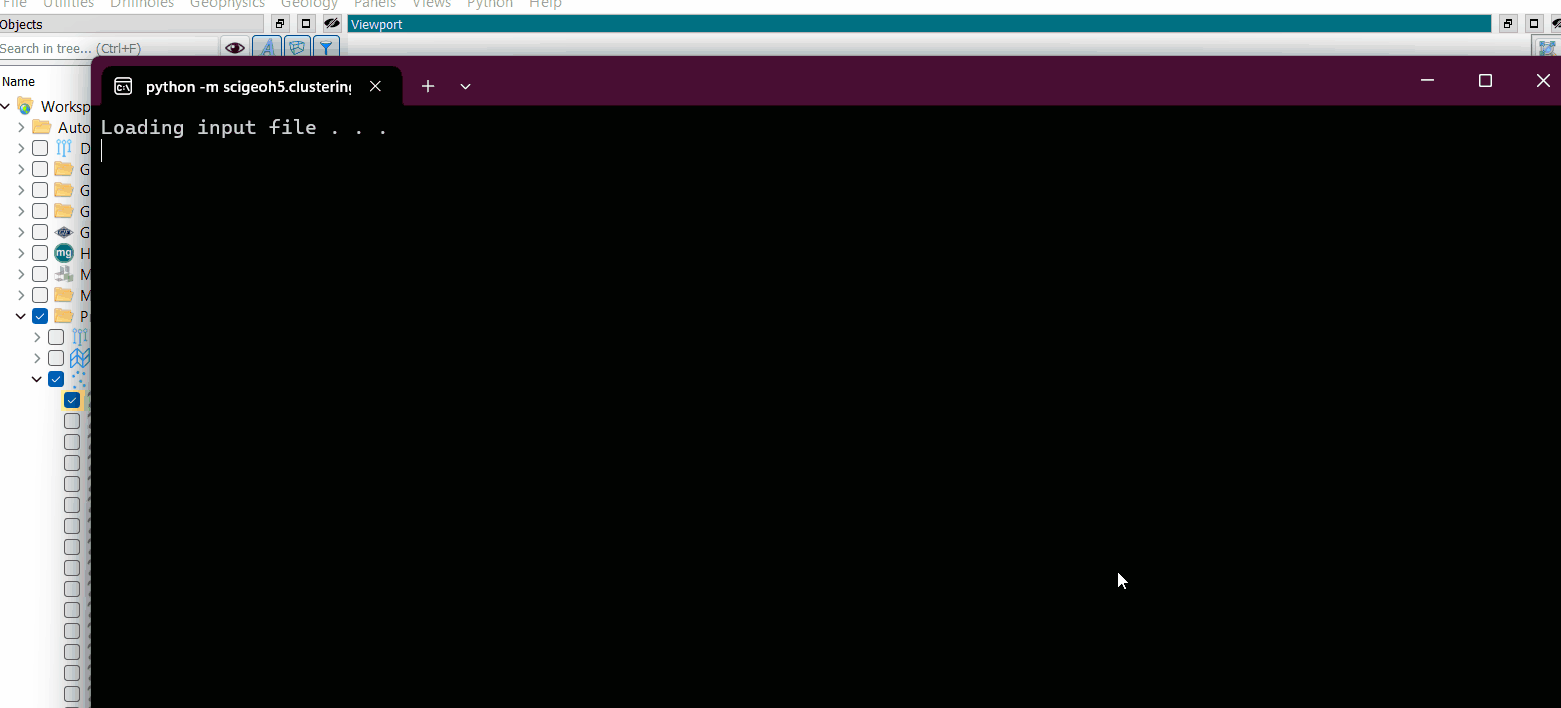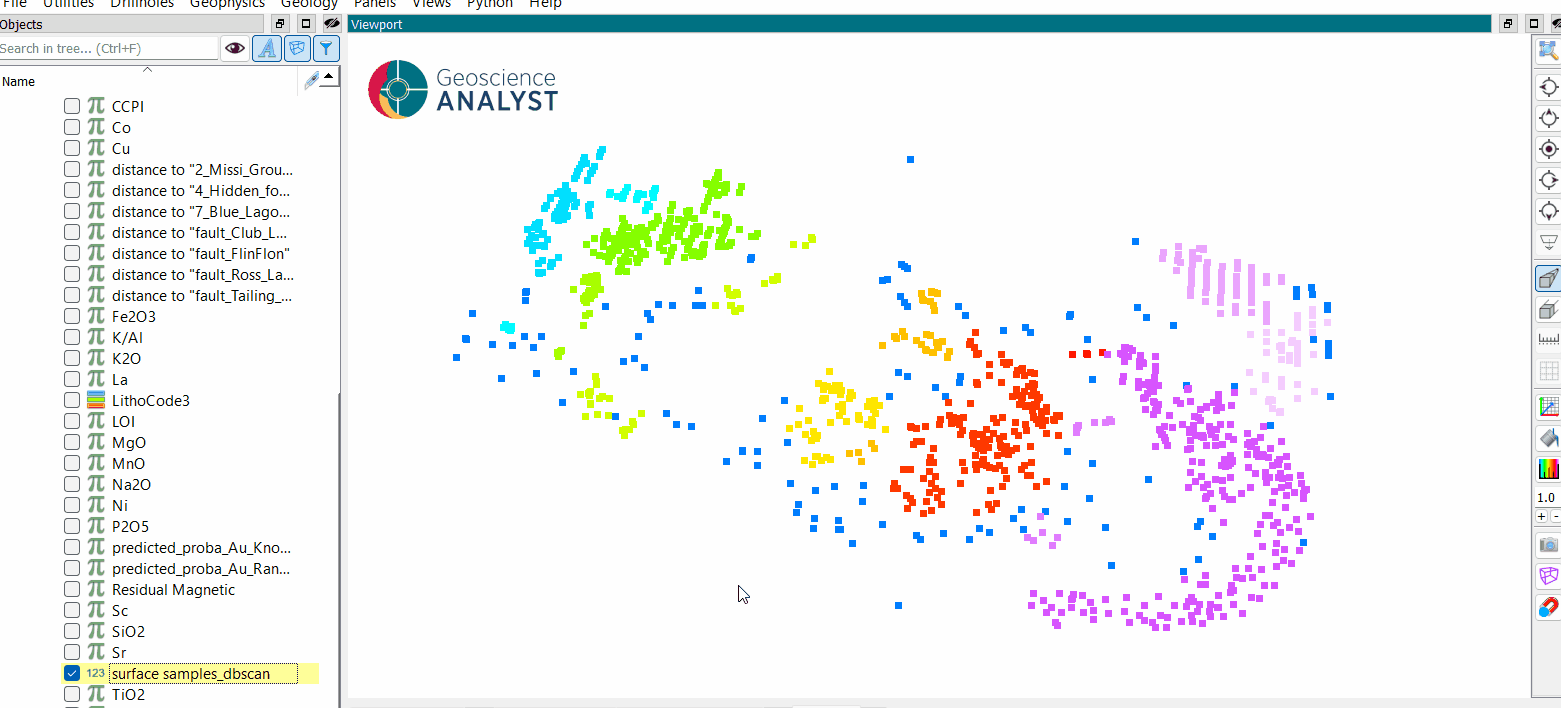
The general parameters controlling the DBSCAN Clustering algorithm are shown in the figure below.
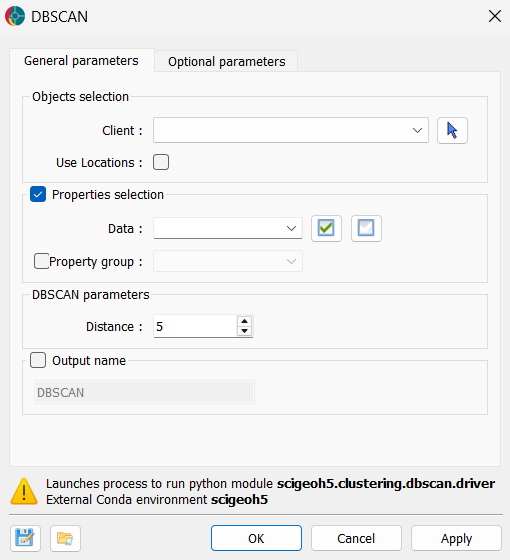
The options are described as follows:
Objects Selection¶
Object: The object containing the data to cluster.
Use Locations: If activated, the spatial locations will be added to the data properties for clustering.
Data: The data properties used for clustering. Users can select multiple
Floatdata properties.Optional. If deactivated, the clustering will be conducted on the spatial locations only.
DBSCAN Parameters¶
Distance (Epsilon): The maximum distance between two points for one to be considered within the neighborhood of the other. This controls how close points must be to be grouped in the same cluster.
Output Names¶
Output Name (optional): The name of the output clustering data. If not specified, the default name will be “DBSCAN”.
Advanced parameters¶
Advanced controls on the DBSCAN Clustering algorithm are available as advanced parameters, as shown in the figure below.
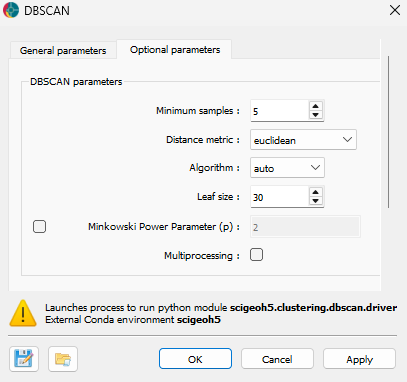
Minimum Samples (default = 5): The minimum number of neighboring points required for a point to be considered a core point and contribute to a cluster.
Distance Metric (default = “euclidean”): Defines how the distance between points is calculated. Available options include:
cityblock: Also known as Manhattan distance, it measures the absolute differences between coordinates.cosine: Measures similarity based on the angle between two vectors.euclidean: The straight-line distance between two points (default).haversine: Computes the great-circle distance between latitude-longitude coordinates.l1: Equivalent to cityblock distance.l2: Equivalent to Euclidean distance.manhattan: Another name for cityblock distance.nan_euclidean: Computes Euclidean distance while handling missing values.
Algorithm (default = “auto”): The method used by the NearestNeighbors module to compute distances between points. Options include:
auto: Selects the best algorithm based on the data.ball_tree: Uses a Ball Tree structure for efficient neighbor searches.kd_tree: Uses a k-dimensional tree for nearest neighbor lookups.brute: Computes distances by directly comparing all points (slowest but exact).
Leaf Size (default = 30): The leaf size parameter used by BallTree or KDTree, affecting query speed and memory usage.
Minkowski Power Parameter (p) (optional, default = 2 if not set): The exponent for the Minkowski distance metric. Common values:
p = 1: Equivalent to cityblock (Manhattan) distance.p = 2: Equivalent to Euclidean distance (default).p > 2: Increases the influence of larger differences.
Multiprocessing (optional): If activated, the clustering process will utilize multiple CPU cores to improve performance (half of available CPUs).
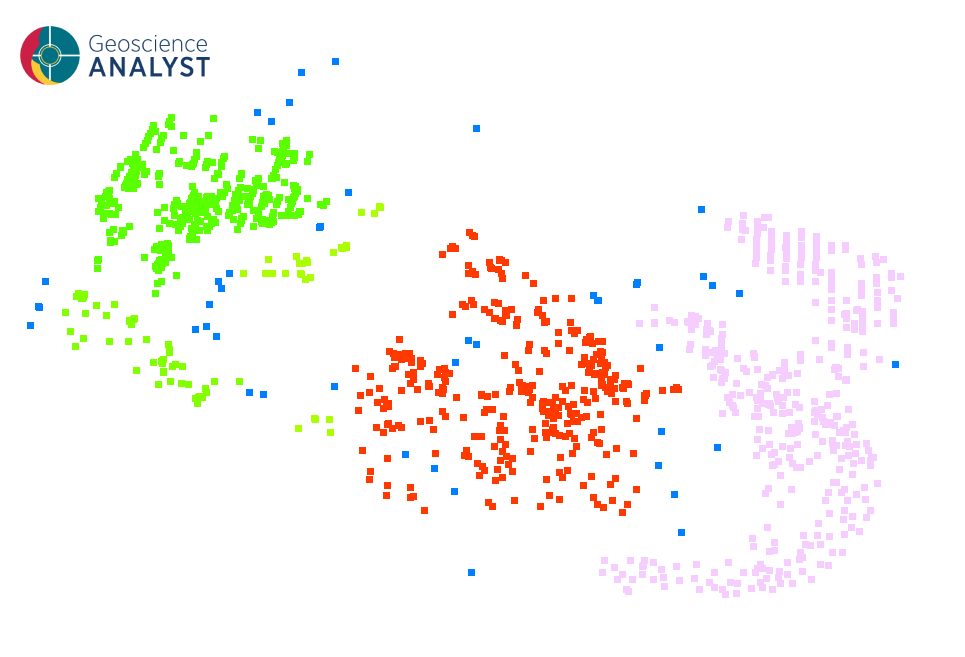
Once the parameters are selected, press OK to run the clustering to obtain a results like the one presented in the first figure.
Tutorial¶
The following animated image presents a tutorial on how to use the DBSCAN application.
Open the application.
Select the object containing the data.
(optional) Use the locations during the clustering.
Select the data to be analyzed.
Define the distance parameter.
Run the application.
Inspect the clustering results.